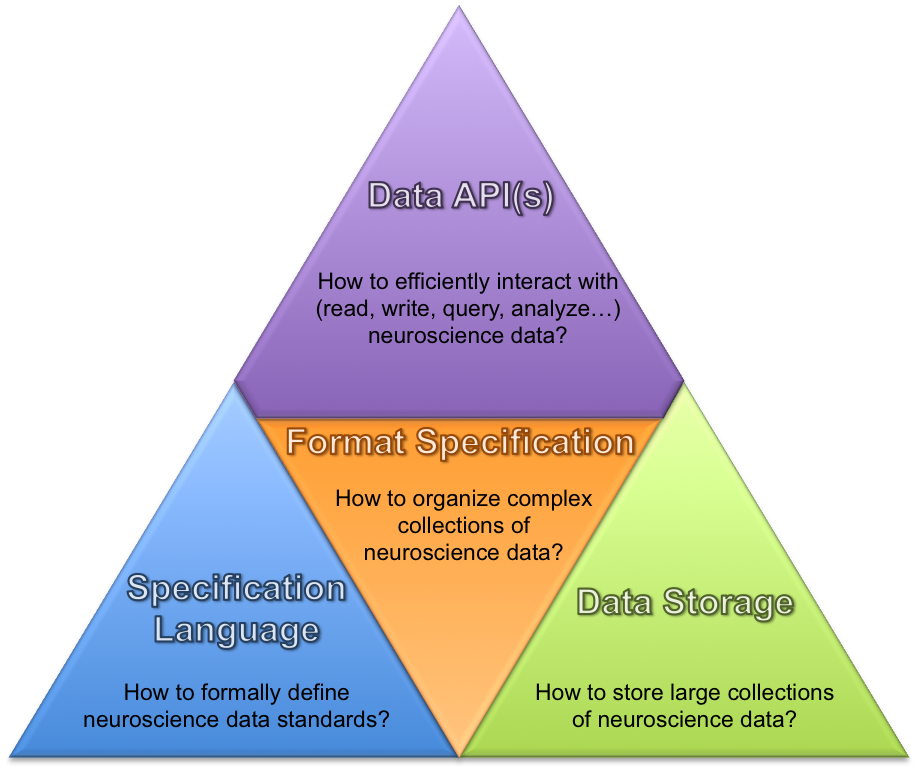
New modular software architecture and APIs to enable users/developers to efficiently interact with the…
Identifying the Brain’s Essential Elements
A database of brain cells, and the new software platform that supports it, may finally reveal the cells’ identities—and supply scientists with a parts list for the brain.
IMAGINE TRYING TO BUILD A MODEL with a blindfold on. It would be hard, if not impossible, to fit all the pieces together properly. Yet that is the challenge brain researchers face everyday because they lack a complete parts list for the brain. Neurons, with names like pyramidal, basket and granule cells, are the brain’s essential elements, but despite more than a century of study, scientists still don’t know how many types there are or the precise characteristics that distinguish one type from another.
The Seattle-based Allen Institute for Brain Science, a not-for-profit that has spent a decade mapping the brain, is aiming to fix that by generating a searchable database that describes thousands of individual cells in the mouse brain. It launched as the Cell Types Database earlier this year, with descriptions of the location, size and electrical signals of 240 cells, as well as detailed three-dimensional reconstructions of their shapes, and will continue to expand.
Read press release, “To Accelerate Pace of Discovery, Neuroscientists and Funders Launch Resource Aimed at Breaking Data Barriers in Brain Research“
Brain researchers worldwide can access this online catalog and use complimentary software tools to conduct virtual experiments, with the ultimate goal of figuring out how to categorize neurons. This is a goal shared by President Obama’s Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative, which has made it a top priority. With such a parts list in hand, modeling the brain and figuring out how it functions becomes a more tractable problem.
Creating the Cell Types Database hasn’t been easy. A team of Allen Institute scientists first had to make measurements of brain cells in a standardized way. They also had to find a way to aggregate different types of data on individual cells, amounting to millions of data points, into a searchable database. That required a new software platform: Neurodata Without Borders:Neurophysiology. It lets neuroscientists capture and share multiple types of data about the brain, and represents a major step toward breaking down the barriers to data sharing in neuroscience.
In a roundtable discussion, The Kavli Foundation spoke to two leaders from the Allen Institute—Christof Koch and Chinh Dang, as well as neuroscientist Kenneth Harris, about why the Cell Types Database and the Neurodata Without Borders:Neurophysiology format matter.
The participants were:
- Chinh Dang – is Chief Administrative Officer at the Allen Institute for Brain Science in Seattle, one of the founding partners of Neurodata Without Borders. She oversees the technologies that enable the Allen Institute’s research and support the Allen Brain Atlases and related resources.
- Kenneth Harris, PhD – is Professor of Quantitative Neuroscience in the Institute of Neurology and the Department of Physiology, Pharmacology and Neuroscience at University College London in the United Kingdom. He studies how neurons organize themselves into functional networks using experimental and theoretical techniques.
- Christof Koch, PhD – is President and Chief Science Officer at the Allen Institute for Brain Science. He is a renowned neuroscientist whose research interests include the neural basis of consciousness.
The following is an edited transcript of their roundtable discussion. The participants have been provided the opportunity to amend or edit their remarks.

THE KAVLI FOUNDATION: Christof or Chinh, why does neuroscience need the Allen Cell Types Database?
CHRISTOF KOCH: We need to know the different components that the brain, particularly the cerebral cortex, is made of. People have spent the last 100 years identifying different brain cells based on their anatomy, electrical properties and, very recently, on the genes that a cell expresses. But that information is piecemeal. It was gathered using different techniques, species and brain regions, and it is published in different papers. We felt there needed to be a repository where all these data types are combined in a standardized way that is accessible to anyone who wants to use it.
KENNETH HARRIS: That’s absolutely right. Neuroscience is only now maturing to the extent that we’ve seen in other fields and undertaking systematic investigations of the brain such as this one.

TKF: Picking up on that, the Cell Types Database is an example of a big-science project involving big data. Chinh, what kinds of challenges did it pose?
CHINH DANG: There were technical, scientific and sociological challenges. For example, the Cell Types Database is the first product of the Allen Institute’s 10-year scientific plan to tackle fundamental problems in neuroscience, so it was the first time that all of the many, many new scientists we’ve hired had to work together. Then there was figuring out of how to scale up the research pipeline in a highly systematic and reproducible manner. The main technology challenge was figuring out how to put the data together so that the neuroscience community could use them because nothing else like this exists.
TKF: Your team has pinpointed single cells in the mouse brain and determined their location, shape and behavior in the form of electrical activity. And soon you will also know what genes each of these cells expresses. How do you expect neuroscientists to use this information?
KOCH: The idea is that people will use it to come up with their own classification systems for neuronal cell types. Using our software, people will also use it to conduct virtual experiments and create models of networks of cells. We have provided models of single neurons that can be downloaded and run in neuroscientists’ own simulators. All of this gives them a very consistent way to model the brain or its components.
DANG: That’s right. We’ve provided the raw data and people can come to ask questions such as “What are the different cell types in this brain region? And how do they relate to one another? We’ve also provided some models that are based on an analysis of the raw, or observational data.

TKF: Let’s talk about the Neurodata Without Borders (NWB) Neurophysiology project. Why has the Allen Institute chosen to use the new NWB data format to support the Cell Types Database?
KOCH: Simply put, we chose it because we helped design it. And we helped design it because the entire field needs a standard so that neuroscience data are interchangeable.
Right now, for even a single technology such as optical imaging there are many different ways to represent and store the data it produces. Things get more complicated when you’re generating more than one kind of data. For example, Karel Svoboda, a neuroscientist from the Howard Hughes Medical Institute (HHMI) and a partner on the Neurodata Without Borders project, visited us here in Seattle not that long ago. Karel does both electrophysiology and two-photon calcium imaging in mice. He says that within his lab, his researchers can’t compare each other’s data because they use very different data standards. That’s really shocking. Imagine how difficult it is to compare data between labs! So it’s of the utmost importance that we come to a standard as have other fields, such as astronomy, so that there’s a way to compare and to contrast data, including metadata. Metadata describe the conditions under which an experiment was performed, for example, what kind of stimulus was used, recording sites, the animal’s behavior. Without them, the raw data are meaningless.
We hope that by adopting the Neurodata Without Borders format, along with a few others, it will spread. Eventually, we hope there will be a phase transition where suddenly the majority of people use it. Then it becomes self-reinforcing because everybody benefits.
TKF: Kenneth, what excited you about the project? Why did you get involved?
HARRIS: The data and particularly the metadata are extraordinarily complex in neuroscience because the experiments involve behavior and you can never predict what the subject is going to do. But for the field to mature, we’ve got to get to the point where we can exchange data between labs and reproduce analyses that other people have done. It’s really long overdue to be honest.
For the field [of neuroscience] to mature, we’ve got to get to the point where we can exchange data between labs and reproduce analyses that other people have done. It’s really long overdue to be honest.”—Kenneth Harris
TKF: Christof, the last time we talked about Neurodata Without Borders, you said that the data format must be convenient for people to use. What kind of feedback have you been getting on the Allen Cell Types Database and the NWB format?
KOCH: Ken, can we consider you an early commentator and enthusiast?
HARRIS: Yes, the NWB format is a very good first step. The great thing is it gets everything—data and metadata—into one place. There will be an evolution to make something the data users and in particular, the builders of analysis tools will find easy to use.
If you think about what’s going to drive adoption of the format in the community, there are two things: The first is the release of large data sets, and that’s exactly what the Allen Institute and the other data providers who are involved with the Neurodata Without Borders:Neurophysiology project are doing. People will have to learn to use the format because that’s how these data sets are formatted. The other thing that I think will drive adoption of the format in smaller labs is if there are killer app analysis tools that someone is going to want to use because they help analyze the data in the most effective way. It’s an ongoing process. I think the next step is talking to the people who develop those tools and seeing what works, how it can be further improved and so on.
KOCH: As Ken said, this is version 1.0. By no means is this a final standard. We’ll have to revise the standard and build additional tools to maximize its usefulness.
TKF: It may be version 1.0, but the Neurodata Without Borders team developed it in about a year. What set this effort apart from other attempts to develop a format?
KOCH: Two things: One is the miracle of deadlines. With Neurodata Without Borders, we gave ourselves one year to establish a data format. We decided we were not going to have an endless academic exercise with huge committees. And two, we haven’t let the better be the enemy of the good. So in less than a year, we have something, but it’s a work in progress. We are currently talking with a number of other players in the field to see if they are interested in adopting the NWB data standard. Whether we can induce a phase transition to where a large fraction of people are using it remains to be seen.
HARRIS: I think there is also the realization in the community that we really need to do this now. That it’s such a high priority for neuroscience. And I think that’s why there’s been a lot of interest in getting it done.
The main technology challenge was figuring out how to put the data together so that the neuroscience community could use them because nothing else like [the Cell Types Database] exists.”—Chinh Dang
TKF: As you’ve mentioned, the biggest challenge that Neurodata Without Borders faces is still in front of you. It’s getting the neuroscience community to adopt a common data format. Aside from making high-value data sets, such as the Allen Cell Types Database available, what else could be done?
DANG: I completely agree with something Ken said earlier: In additional to standardizing large data sets, it’s just as important to have data acquisition tools that write out the data in that format and analysis tool that make use of the data format. We’ve done the first step in this first year to get a schema of an NWB that can support both electrophysiology and optophysiology data, but I think that the next phase of the NWB project should be focusing on how to work with acquisition systems and analysis packages that are already out there.
TKF: Are they interested?
HARRIS: Absolutely. Everyone realizes this has to happen.
KOCH: And the funders of scientific research could also do their part by requiring that people make their data public and also that they use this or some other common data standard as part of that. If the National Institutes of Health, which funds the vast majority of neuroscience research in this country, were to mandate the use of such a standard, it would happen very quickly.
TKF: What’s next for the Allen Cell Types Database?
KOCH: We’re working on a transcriptional analysis of the same cells, which means what genes it expresses. So for each cell, we will be able to visualize where it is in the mouse visual cortex, how it looks, what its electrical behavior is, and also the genes that it expresses.
Secondly, we’re working on a cell types database for the human brain with the help of neurosurgical clinics here in the Pacific Northwest. We receive pieces of brain about the size of a sugar cube that 20 minutes ago were a part of someone’s brain. A neurosurgeon had to remove them from a particular part of a patient’s cerebral cortex to get to an underlying tumor or epileptic foci. Now, rather than being burned as medical waste, the piece of brain can be used to do the same experiments we have done in mice. So, for example, we can record the electrical activity of different cells. The remarkable thing is that just a half hour ago the cells we’re studying were part of somebody’s brain and maybe part of a circuit that contained the memory of their first kiss.
It turns out to be much easier to record from these human brain cells than from those of mice. They’re bigger and they’re much more resistant to stress. So we can typically record from these human neurons for two or three days. With a mouse, you typically have six or eight hours.
So we expect to release much more mouse data, including gene expression data, and then the human data, all using the Neurodata Without Borders format.
We’re working on a cell types database for the human brain with the help of neurosurgical clinics here in the Pacific Northwest…. The remarkable thing is that just a half hour ago the cells we’re studying were part of somebody’s brain and maybe part of a circuit that contained the memory of their first kiss.”—Christof Koch
TKF: Scientists tend to guard their data because it’s the heart of their research programs and, ultimately, of their success. The Allen Institute takes the opposite stance. Sharing data, tools and knowledge with the neuroscience community is at the heart of your mandate. Chinh, what kind of impact is your open-science policy having?
DANG: Well, there are quantitative as well as qualitative ways of measuring that. We have more than 50,000 users coming to our site every month to use our databases, which amounts to more than 2 million visitors to date. We also know that many, many scientists around the world use our data to publish on. In fact, I know of a few different graduate students who have built their entire PhDs on mining and using our data. On the qualitative side of things, Ken may be a better person to address this. But I hear of people all the time who say, “Oh, yeah, I take a look at your data to get ideas of how to design my experiments,” but they would not be citing us.
HARRIS: So from my perspective as a user, the Allen Brain Atlas, which maps gene expression throughout the adult mouse brain, changed absolutely everything about the way we do neuroscience. It used to be that if someone mentioned the name of a gene, you’d think, “Oh, well I don’t know what that is.” But now the first thing you do is go to the Allen Brain Atlas to see where it’s expressed in the brain. That gives you an intuitive understanding of what it might be doing. Or if you’re interested in a particular layer of the cerebral cortex, you’ll look at that layer in the Atlas to see what genes are expressed there. These are questions that were just completely hopeless before. As Chinh says, you can’t quantify its impact simply through citations. It’s just become deeply ingrained in the community. I expect that the same thing will happen with the Cell Types Database.
TKF: As the Allen and other neuroscientists release more data that uses the format, what do you hope the impact will be?
HARRIS: I would hope this encourages some sort of a cultural change in the field. At the moment, you’re either an experimentalist who collects the data or you’re a modeler who uses the data to build computational models. But there’s a third career path, which is to be someone that analyzes existing data and finds new patterns it. By analogy, the astronomer Tycho Brahe, from way back in history, collected a vast amount of data on the motions of the planets. But he didn’t quite understand them, or maybe he wasn’t interested in making a mathematical model from them. Instead, it was Johannes Kepler who found the patterns in the data. There could be a similar career path in the neurosciences in which someone analyzes experimental data and doesn’t even need to do experiments. I think when the data are out there and in a standard format, that will become a possibility.
KOCH: Incidentally, Ken, Tycho Brahe’s work is one of the first examples of big-science discoveries. He may not have been the first, but he was probably the most accomplished person who cataloged the position of the stars before the advent of the telescope. And that work led him to one of the most important discoveries in the history of astronomy—what we now call SN 1572, the stellar nova that today we call supernova. In the late 1500s, he observed this new star and had evidence to show that Aristotle’s view of the static sky was wrong; that there was actually change in the cosmos, that new starts could be born. That was only partly because he systematically mapped the position of stars, and then found, “Hey, that star wasn’t there before.” I think that’s one of the first and most beautiful instances of somebody who catalogs things and thereby makes a fundamental discovery.
—Lindsay Borthwick