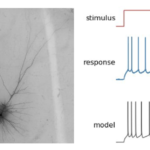
The DANDI archive is the best place to find published datasets in NWB 2.0. Below, we feature several datasets as representative examples across the community. For the most up-to-date information, to search across datasets, and to find new datasets, visit the DANDI website.
Extracellular electrophysiology
Visual Coding Neuropixels Data, Allen Institute for Brain Science
The Allen Institute for Brain Science has released its first — and the world’s largest — dataset of electrical brain activity gathered using Neuropixels, a new high-resolution silicon probe that can read out activity from hundreds of neurons simultaneously. This initial data release consists of 58 experiments. Each experiment contains data from up to six Neuropixels probes recording in cortex, hippocampus, and thalamus. Data for each experiment is conveniently packaged in Neurodata Without Borders (NWB) files that can be downloaded via the Allen SDK. For further details see the news release by the Allen Institute as well as the Visual Coding – Neuropixels website.
Human single-neuron activity during a declarative memory task, Rutishauser Lab, Cedars Sinai
DOWNLOAD on DANDI. Publication. Conversion code.
Neuropixel recordings during visual decision making, Steinmetz Lab
Neuropixel probes recording extracellular electrophysiology simultaneously from a variety of brain regions in mice engaged and a visual decision task.
DOWNLOAD on DANDI. Publication. Conversion code.
Hippocampal electrophysiology during spatial navigation, Buzsáki Lab, NYU
Electrophysiology recordings from hippocampal regions of mice in theta-maze and in open exploration, from Senzai, Yuta, and György Buzsáki. “Physiological properties and behavioral correlates of hippocampal granule cells and mossy cells.” Neuron.
Intracellular electrophysiology
Example of metadata extension, Blue Brain Project
This dataset shows the new metadata features offered in ndx-icephys-meta that provide a standardize structure for expressing intracellular electrophysiology experimental design such as sweeps and conditions.
Electrophysiology data from thalamic and cortical neurons during somatosensation, Svoboda Lab, Janelia
Intracellular and extracellular electrophysiology recordings performed on mouse barrel cortex and ventral posterolateral nucleus (vpm) in whisker-based object locating task.
Optical Physiology
Calcium imaging during decision making, Anne Churchland Lab, Cold Spring Harbor Laboratory
Calcium imaging dataset accompanying the paper:
Farzaneh Najafi, Gamaleldin F Elsayed, Robin Cao, Eftychios Pnevmatikakis, Peter E Latham, John Cunningham, Anne K Churchland. “Excitatory and inhibitory subnetworks are equally selective during decision-making and emerge simultaneously during learning” bioRxiv (2018): 354340.
DOWNLOAD on DANDI. python demo. MATLAB demo.
A Map of Anticipatory Activity in Mouse Motor Cortex, Svoboda Lab, Janelia
Data from “A Map of Anticipatory Activity in Mouse Motor Cortex” Chen et al. Neuron 2017
Allen Cell Types Database
There are 86 billion neurons in the adult human brain, and like snowflakes, no two are exactly the same. Brain cells are found in a dizzying array of intricate shapes, have different electrical patterns and functions, and express different genes. Sorting these cells into “types” is an enormous and complex challenge, but before we can hope to understand our brains, we need to create a rich list of its building blocks.
The Allen Cell Types Database is a new tool to help scientists create that list, and to understand what makes one type of cell different from another. “Identifying neuronal cell types is essential to unraveling the mystery of how the brain processes information and gives rise to perception, memory and consciousness,” says Christof Koch, President and Chief Scientific Officer of the Allen Institute for Brain Science. “This is the first resource of its kind to bring together multiple types of data—shape, position in the brain and electrical activity—in a single searchable database anywhere on the planet.”
Data in the Allen Cell Types Database is organized in an alpha version of the NWB format and will be updated in later releases.
Overview of the Allen Human Brain Atlas
Cell Feature Filters

Electrophysiology Data Summary
NWB:N 1.0 Datasets
NWB:N data sets have been published in the newer NWB 2.0 standard and the older NWB 1.0 version.
For an updated list of NWB:N 2.0 data sets, see here: https://neurodatawithoutborders.github.io/exampledata. These data sets can be read by the latest versions of PyNWB and MatNWB.
The majority of NWB:N 1.0 data sets are hosted at CRCNS.org. CRCNS (Collaborative Research in Computational Neuroscience) is a joint program of NSF and NIH that supports integration of theoretical and experimental neuroscience through collaborative research projects.
The following data sets represent the primary use cases for the development of the NWB format:
- Rat hippocampus, contributed by György Buzsáki, New York University (CRCNS hc-3 data set)
- Mouse slice electrophysiology, contributed by the Allen Institute (CRCNS pvc-6 data set)
- Electrophysiological recordings from rat barrel cortex, contributed by Karel Svoboda, Janelia Research Campus (HHMI) (CRCNS alm-1 data set)
- Calcium imaging of rat somatosensory cortex, contributed by Karel Svoboda, Janelia Research Campus (HHMI) (CRCNS ssc-1 data set)
- Retina, contributed by Markus Meister at the California Institute of Technology (CRCNS ret-1 data set)
This list is no longer kept updated. New users of NWB:N are recommended to use datasets published in NWB 2.0 (see above) as a reference.
Disclaimer:
Reference herein to any specific data, product, process, or service by its trade name, trademark, manufacturer, or otherwise, does not constitute or imply its endorsement, recommendation, or favoring by the NWB team, United States Government or any agency thereof, The Regents of the University of California, or the Kavli foundation. Use of the Neurodata Without Borders name for endorsements is prohibited.