NWB is involved in several working groups to curate the core NWB standard, define ontologies, and integrate with other community standards and tools. Please contact us if you are interested in learning more or getting involved with one of these groups.
Genotype representation
The aim of this working group is to define an NWB extension to enable scientists to systematically report the genotype of the transgenic animal used in their physiological experiments. Cell-type-specific recordings are often based on transgenic mice or rats. Accordingly mature infrastructure, such as public repositories and centralized databases, exist to support the scientific community in the use of these tools. We are extending the NWB standard to integrate and leverage these resources for storing relevant metadata on the genotypes and genetic backgrounds of individual mice, linking neurophysiological data to the databases and literature that characterize the experimental animals. [NDX Proposal]
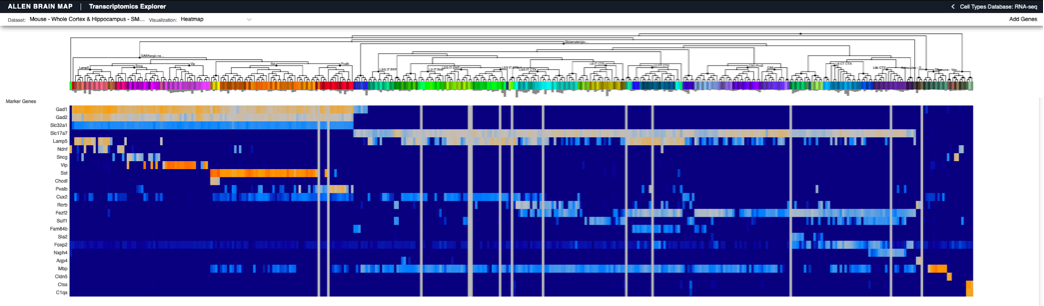
Spatial coordinates representation
Reporting of spatial coordinates within the brain is a complex problem, requiring registration between data collected with multiple recording modalities (imaging, probes) and across brain preparations (in vivo whole brain, slice) with common reference atlases and controlled vocabularies. The focus of this working group is to create an NWB extension to capture spatial coordinate systems, spatial mappings between two systems, and the representation of objects (e.g. cells, probes, injections, and optogenetic stimuli) within a spatial coordinate system. This type of standardization will facilitate the programmatic integration and comparison of data represented in NWB with other datasets that have been mapped to the same coordinate system. [NDX Proposal]
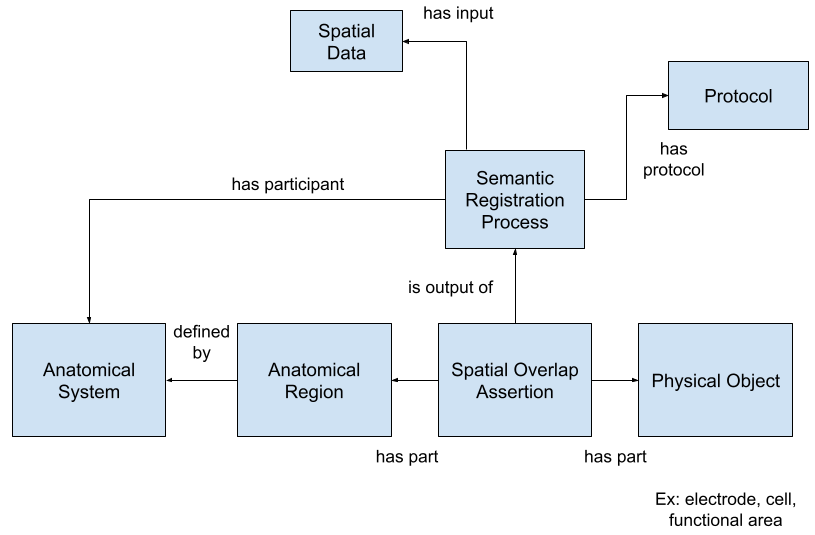
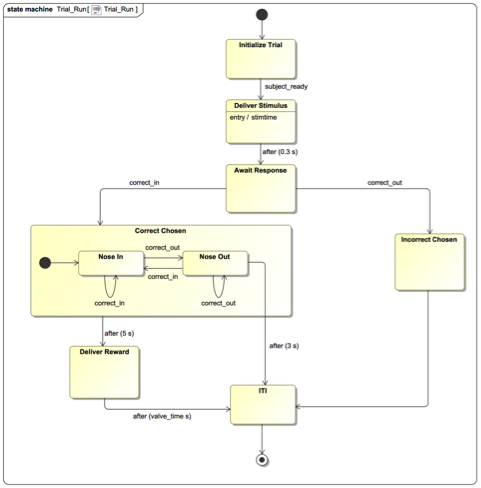
Behavioral task representation
Behavioral tasks are an increasingly important component in neurophysiology experiments, and can have considerably more complex designs than passive sensory studies. There is currently no capability in NWB for storing the data and metadata that specify the control logic and behavioral contingencies that determine the flow of a behavioral session. The goal of this working group is to incorporate new features for improving descriptions of behavioral tasks within NWB. In this work, we are collaborating with the Kepecs lab at Washington University who are building a standard markup language (BEADL) to build state machine descriptions of task logic and control flow within a trial and across trials in a session, including tools for visualization, construction, and emulation of tasks from these machine-readable descriptions. [NDX Proposal]
External Working Groups
NWB team members also participate in numerous external working groups. While the charge of external working groups are often broader than just NWB, they all involve components that are relevant to NWB and commonly use NWB or are creating a Neurodata Extension (NDX) as part of their charge.
INCF Working Group on standardization of a directory structure containing neurophysiology data
The objective of this project is to put together a set of specifications and tools that would allow the standardization of a directory structure containing experimental data recorded using animal models in neuroscience. The aim is to capitalize on the success of BIDS for human neuroimaging data, while retaining the specificities of datasets obtained in animal models. Such a standardized data structure will facilitate obtaining reproducible research and data sharing following the FAIR principles. In addition to data structures, the working group will discuss what experimental metadata is essential, and which formats and structures are appropriate for different metadata types. [Working Group Page]
INCF Electrophysiology Stimulation Ontology Working Group
Stimulus protocols are an essential component of neurophysiological experiments, and while the possibilities for stimulus protocols are endless, most studies still draw from a small set of commonly used protocols with stereotyped stimulus classes that allow standard analyses to be performed and results compared with other datasets. The aim of this working group is to define an extension for NWB that enables the storing of 1-dimensional stimulus waveform data and metadata with references to an ontology for standardization. This will facilitate comparing and identifying similar protocols across datasets and development of shared analysis scripts for quantitative feature extraction. For the first set of use cases, the group focused on intracellular electrophysiology, as most protocols use very similar protocols with waveforms that have stereotyped profiles (e.g., long steps, ramps, chirps). This working group will also draft recommendations for representation in NWB of parameterized stimuli beyond 1-D waveforms. [Working Group Page]
Standardization of data/file formats for simulation outputs in computational neuroscience
Both neuromorphic and HPC simulations can consume and generate data at very high rates. File formats and associated software libraries for such data need to be able to accommodate this requirement. In recent years a number of projects have been set up which address this (e.g. Brion, SONATA, SIONlib/NEST) , and the focus of this working group is to coordinate efforts between related projects to improve interoperability and make it easier to develop tools for visualization and analysis of such data. Tasks for this working group include compiling a list of existing formats, defining evaluation criteria and evaluating the different formats, and identifying avenues for improving interoperability. [Source][NDX]
Past Working Groups
Hierarchical structures for intracellular electrophysiology
Goal: In a neurophysiological experiment, individual recordings from a session are typically related by experimental design or condition, e.g. multiple time series that constitute a simultaneously recorded sweep or multiple recordings that result from repetitions of the same stimulus. The goal of this working group has been to define a structure for saving these data in the NWB format such that the structure of an experiment is clear and facilitates efficient browsing and retrieval of the data for users. Using intracellular electrophysiology as a first use case, the working group implemented a hierarchical table structure that facilitates grouping of data and metadata by experimental condition.
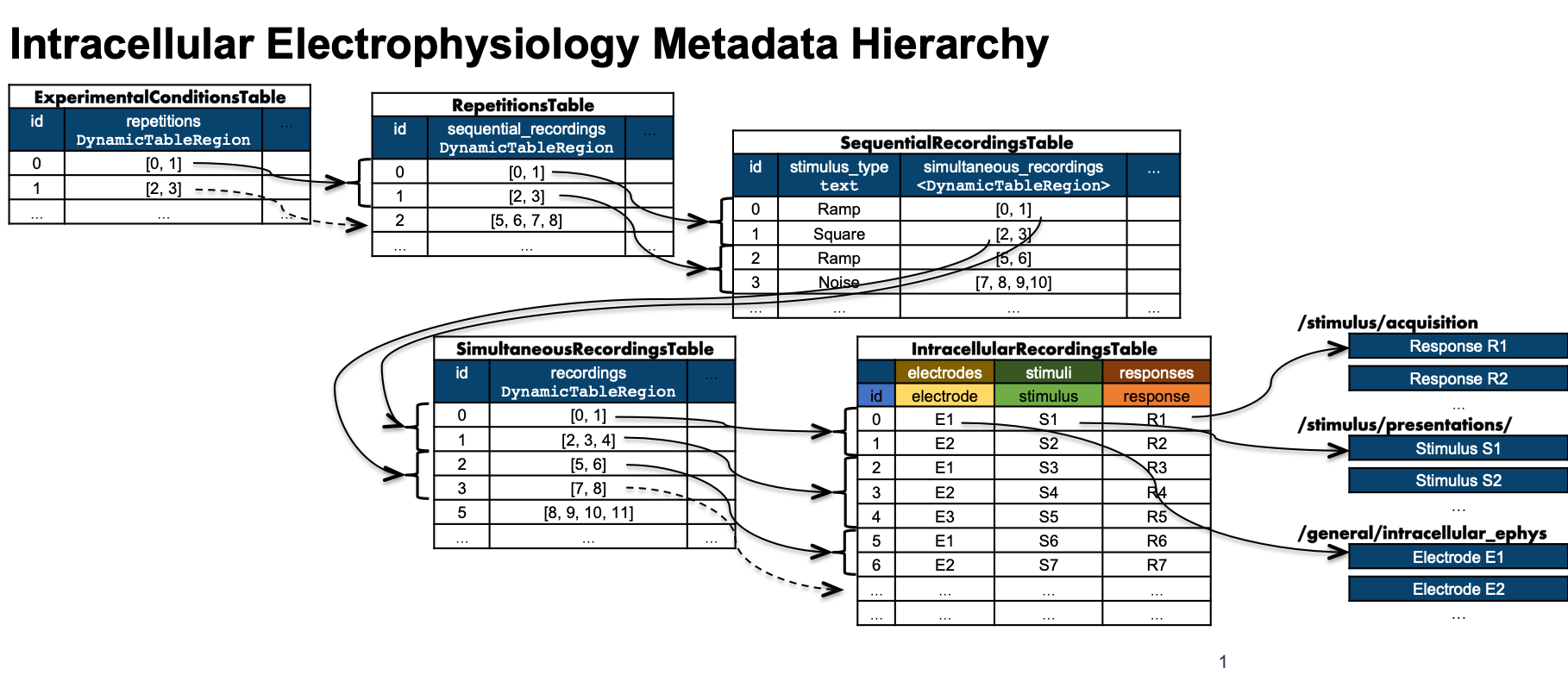
Outcomes: The working group drafted an extension for the NWB schema that implements these tables for testing and evaluation of the proposed changes (see [Source][NDX][NDX Proposal][YouTube]). The proposed extension underwent community review and has been merged with NWB after completion of the review. The new structures developed by this working group have been released as part of NWB Schema version 2.4 and corresponding PyNWB 2.0 and MatNWB 2.4.0.0. These new features are documented in the Intracellular electrophysiology tutorial as part o PyNWB and AlignedDynamicTable tutorial and AlignedDynmaicTable schema as part of HDMF.
